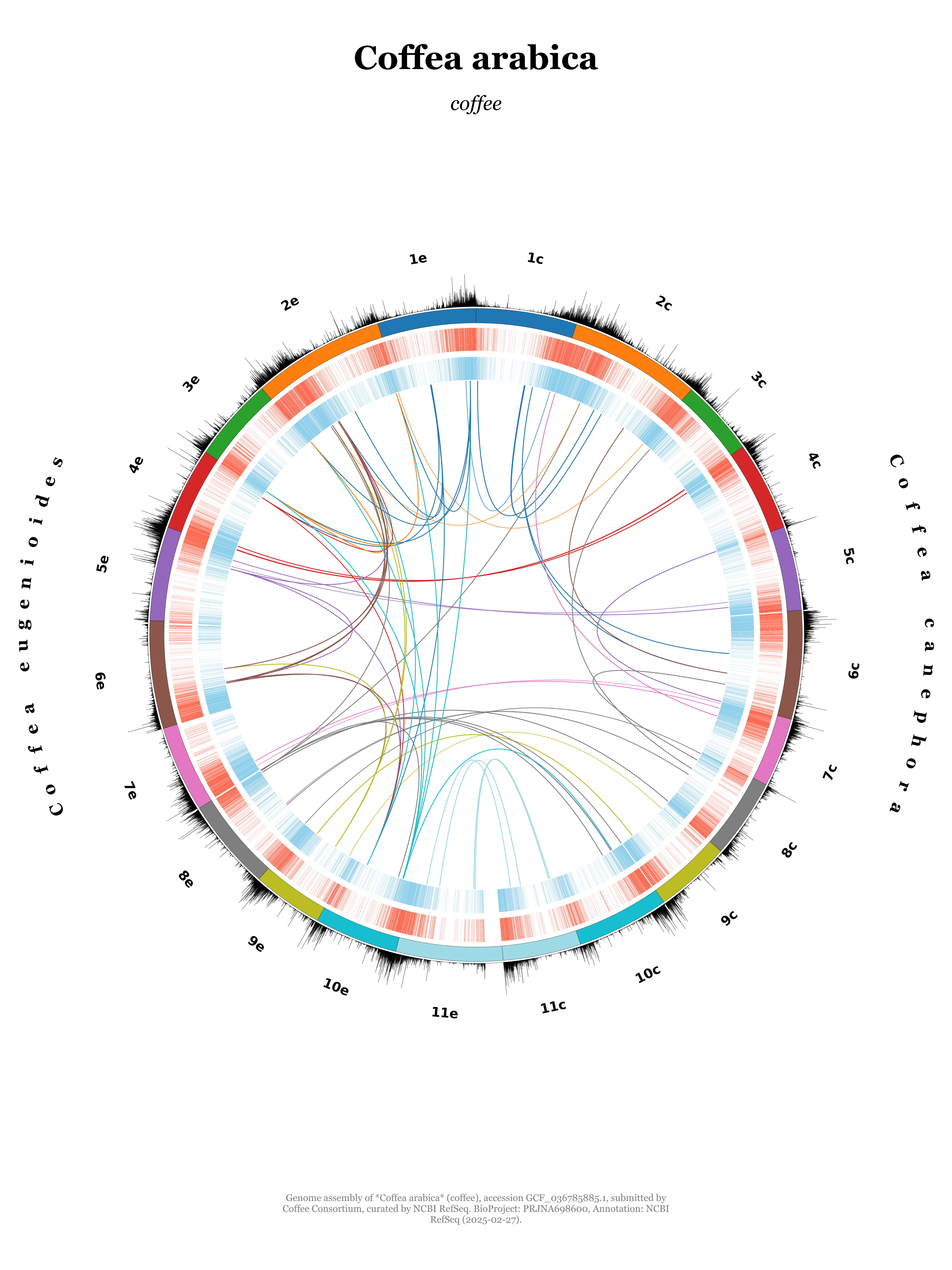
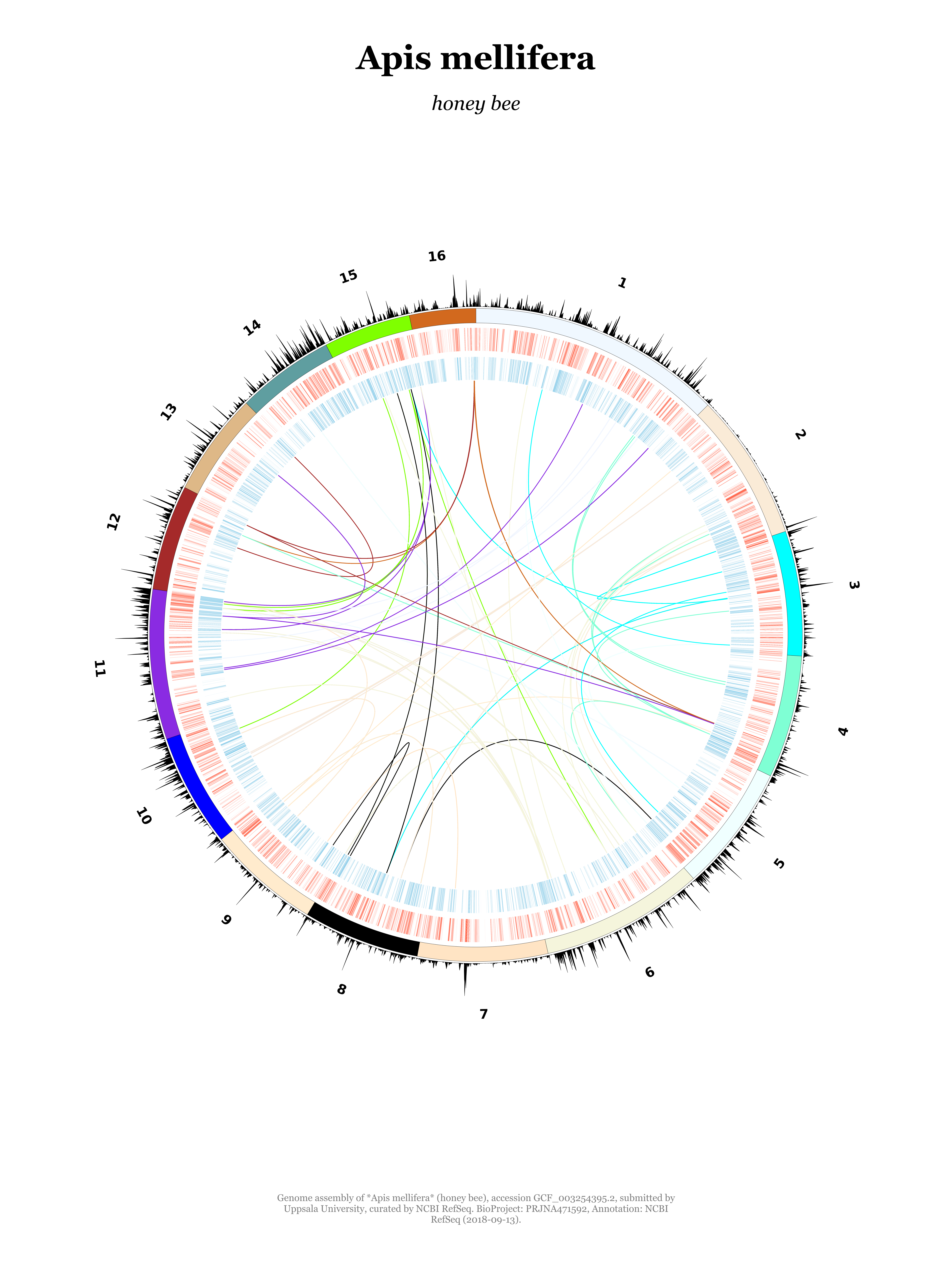
Circos Plot Workflow
This project showcases a Python-based workflow for automating genomic data retrieval and visualization. Using NCBI assembly accession numbers, the pipeline downloads genome annotations (GFF3, FASTA, and metadata), builds a BLAST protein database, performs self-BLAST analyses, and extracts key genomic metrics such as coding region density, gene identity scores, and gene length distributions.
The processed data are then rendered into publication-ready Circos plots, dynamically generated for any species dataset, revealing structural and functional genome patterns.
Project Summary
I developed a Python workflow that automates the retrieval and visualization of genomic data from NCBI Datasets. Using assembly accession numbers, the pipeline downloads GFF and FASTA files along with assembly metadata, builds a local BLAST protein database, and performs a self-BLAST against the assembly’s protein.faa. It then extracts coding regions, top gene identity scores, gene lengths, densities, and replicate mappings to generate publication-ready Circos plots that summarize genomic relationships and structure.
Tools & Skills
- Programming: Python (data processing, automation, and visualization)
- Bioinformatics Tools: NCBI Datasets CLI, BLAST+ (makeblastdb, blastp)
- Data Formats: GFF3, FASTA, JSONL (assembly and sequence reports)
- Libraries: pycirclize, matplotlib, pandas, numpy, json, subprocess, os
- Visualization: Circos-style genome plots, CDS density, BLAST link mapping
- Workflow Design: Reproducible data pipeline with dynamic species handling and figure generation
- Command-Line Integration: Automating external bioinformatics tools within Python
Example Visualizations